Next Generation Sequencing (NGS)
Sanger sequencing-based on Capillary Electrophoresis was the workhorse for the completion of the Human Genome Project that took more than 10 years.
Next-Generation Sequencing (NGS) is a widely used technology in many fields, including genomics, epigenetics, transcriptome, etc. NGS is not only used in basic research but also applied for clinical diagnostics such as cancer screening and drug development. NGS technologies can be crudely categorized by the sequencing read length into short-read sequencing by synthesis, ion semiconductor, sequencing by ligation, and pyrosequencing. And long sequencing reads are much shorter, comprised of two: single-molecule real-time sequencing and nanopore sequencing.
Currently, many commercial NGS platforms have been developed which are based on different principles (e.g., Roche 454FLX, Illumina Genome Sequencer, Applied Biosystems / ABI SOLiD Sequencer). No matter what kind of platform, library preparation is the most critical step in NGS workflow. The major idea of sequencing library preparation is producing adaptor-included DNA fragments with specific size ranges. Therefore, monitoring sample size and quality from sample preparation to sequencing library validation become quite important.
The fully automated Qsep series Bio-Fragment Analyzers (Qsep1, Qsep100 & Qsep400) are able to completely cover the QC protocol from upstream of genomic DNA, total RNA to the final step of the DNA library. With ready to use gel cartridge, users can set up the instrument in 1 minute and results within 2-5 minutes. The data can also be released by batch or individually with fully detailed information. The data demonstrate that the specified detection limit of 50 pg library sample can easily be reached by Qsep instruments.
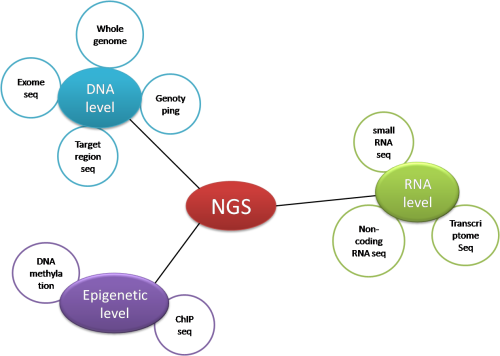
Figure 1. Applications of NGS
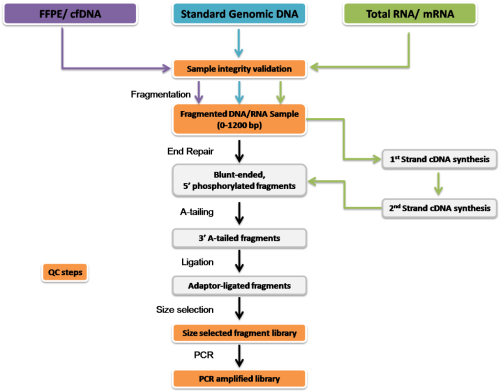
Figure 2. Workflow of Sequencing library construction including many steps of quality control which play an important role to make sure of successful analysis
Qsep1, Qsep100, and Qsep400 systems provide an efficient methodology to fully cover the serial QC steps, which also can match all the NGS instrument workflow.
Analytic steps:
1. Insert gel-cartridge and place the samples
2. Select the proper methods (See the table below for details.)
3. Click "Run"
4. Check and analyze results
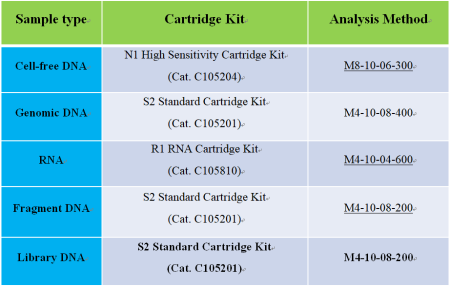
1. CGE analysis:
To evaluate the suitability of the system, the degradation level of smeared genomic DNA samples was analyzed to assess their quality. The ability to rapidly distinguish between intact and degraded genomic DNA samples was crucial for success in sequencing (Figures 3a & 3b).
RQN (RNA Quality Number) value determines the integrity of RNA (Figure 4). By the use of high detection sensitivity N1 cartridge Low amounts of cfDNA (yields less than 5 pg/μl), is directly detectable by the Qsep100 instrument.
2. Results:
Qsep100 system is a high-quality control platform for NGS, which provides the easiest & simplest solution for monitoring each stage of library preparation to ensure success in sequencing. The fully automated system completely covers the QC protocol from upstream of genomic DNA, total RNA to the final step of the DNA library. With ready to use gel cartridge, users can set up the instrument in 1 minute, and get results within 2-5 minutes. The data can also be released by batch or individually with fully detailed information.
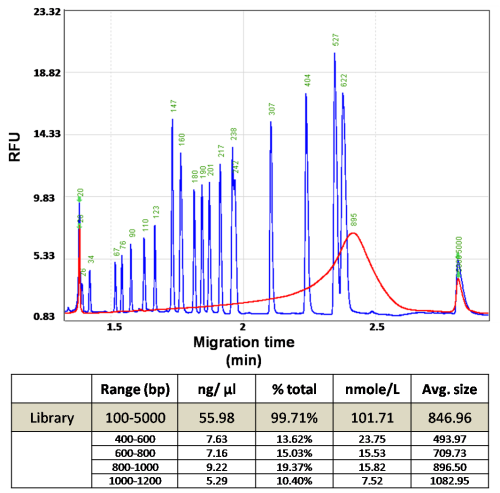
Figure 3a. Quality control of fragmented DNA. DNA is sheared into small fragments with the average size of 895 bp.
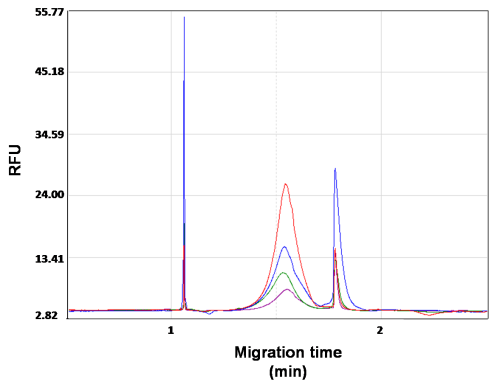
Figure 3b. High sensitivity result of serially diluted library sample. The average size of the DNA library is 515 bp. Library concentrations are from 0.33 ng/μl to 0.07 ng/μl.
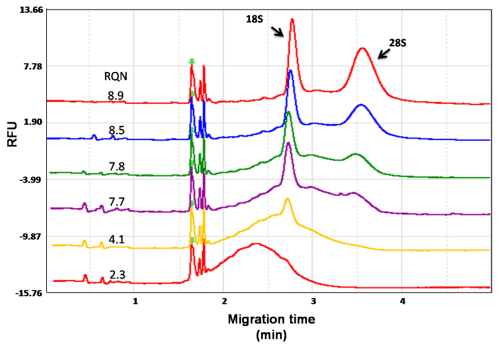
Figure 4. Monitoring integrity of total RNA is a typical QC step for downstream experiments, such as RNA-seq. Samples range are from intact (RQN 8.9) to degraded (RQN 2.3). RQN (RNA Quality Number) value determines the integrity of RNA.
