Chromatin Immunoprecipitation (ChIP)
Chromatin immunoprecipitation (ChIP) refers to a procedure used to investigate the interaction between proteins and DNA in the cell. ChIP is a method used to determine the location of genome binding sites for a specific protein of interest, giving invaluable insights into the regulation of gene expression. ChIP involved the selective enrichment of a chromatin fraction containing a specific antigen. Antibodies that recognize a specific protein or protein modification are used to determine the relative abundance of the antigen at specific loci.
Techniques commonly used to characterize protein–DNA interactions include electrophoretic mobility shift assay (EMSA), DNAse footprinting, and chromatin immunoprecipitation (ChIP) assays. Though EMSA is a rapid and sensitive method to detect protein–DNA interactions, despite its advantages, it has limitations like the samples are not in chemical equilibrium during the electrophoresis step and also many complexes are significantly more stable in the gel than at free solution. ChIP assays minimize these limitations and provide an efficient tool to determine these protein–DNA interactions occurring in vivo, by immunoprecipitating chromatin with specific antibodies. These assays provide unbiased observations into the chromatin changes occurring in response to extracellular signals or during differentiation and development. Although transcription factor recruitment to the promoter is very dynamic, ChIP assays provide an efficient means to study these events.
Qsep1 and Qsep100 Bio-Fragment Analyzers are simple-to-use CGE instruments for the QC testing of Chromatin.
Test Setup:
• Cartridge: S2-O-170612-1
• Alignment marker: 20-15Kb
• Method: M4-10-04-600
• Dilution factor: 2 times diluted by 1x dilution buffer
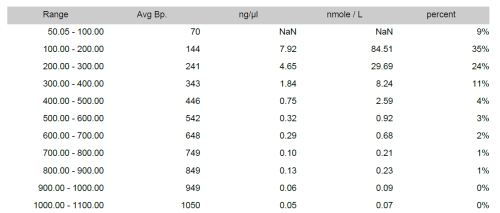
• Marker calculation: From 100 bp to 10Kb
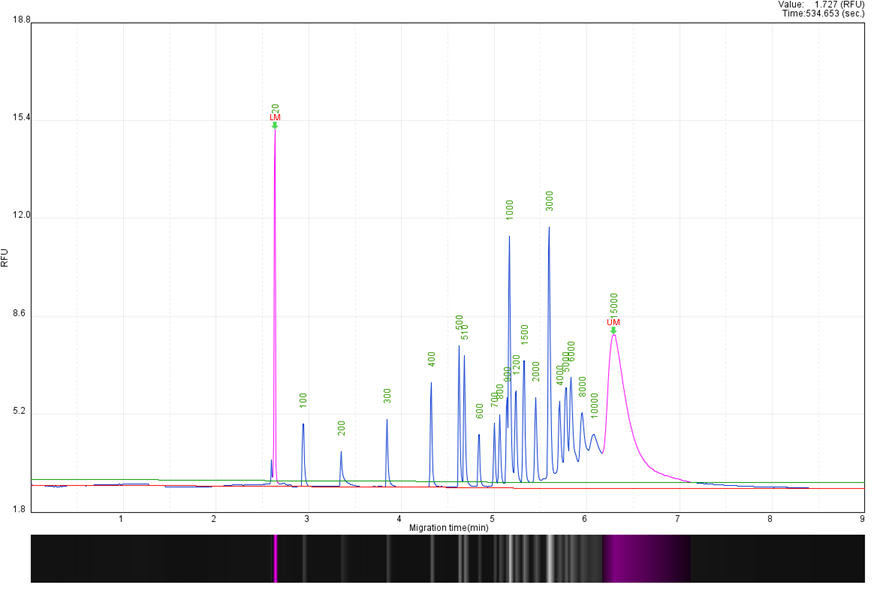
Figure 1. DNA Ladder with 20 + 15Kb Alignment Marker
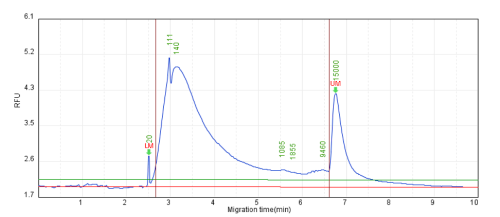
Figure 2. Chromatin detection using the 20+15Kb Alignment Marker